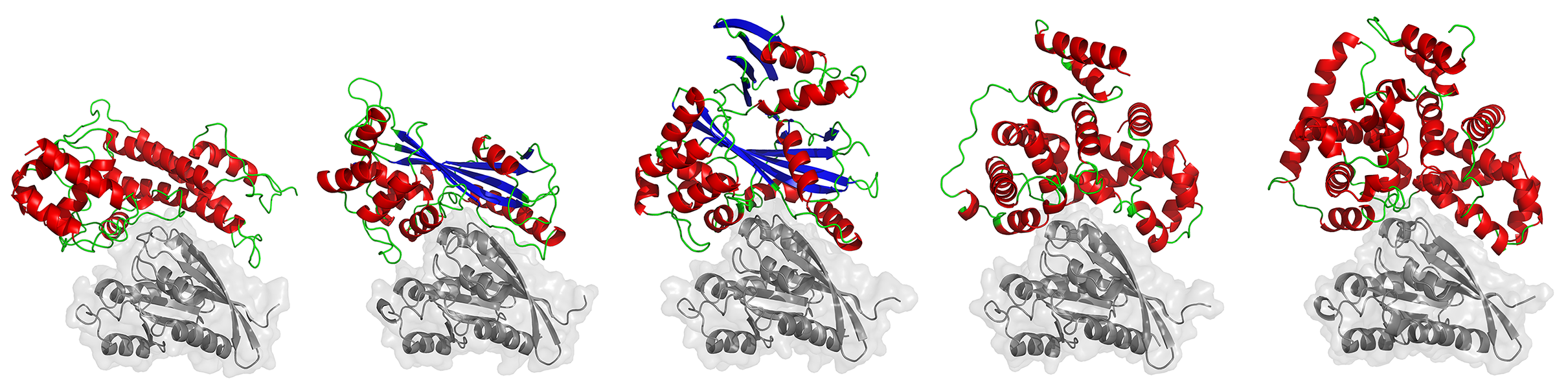
Inbal Mermershtian from the Kosloff lab used a computational structural bioinformatics pipeline that requires hundreds of cores working in parallel for accurate atomic-level calculations of protein-protein interactions - analyzing multiple proteins 3D complexes quantitatively.
In this project Inbal focused on the small G-proteins Rabs, comparing their modulation by host GAP proteins (that turn Rab switches “off”) with the modulation by pathogen GAP proteins. She use a computational method developed in the Kosloff lab that applies structure-based electrostatic energy calculations to pinpoint structural determinants that are critical for fine-tuning protein-protein interaction specificity.
Inbal's analysis uncovers a convergent structural basis for Rab recognition and modulation by eukaryotic and bacterial pathogens, and highlighted key specificity-determining interactions that might be used to target such pathogens with new antibiotics. More generally, this study presents a fascinating case-study of how similar interaction specificity can be achieved by structurally-dissimilar proteins.