Have you ever failed in something and yet felt very proud? This project is the ultimate example for that. This is the Ph.D. project of Felix Glinka in the Privman lab , which is a collaboration with the lab of Deborah Gordon in Stanford. Deborah's lab has been studying this unique and interesting population of harvester ants in the Arizona desert for almost 40 years. She sent Felix 2 ant samples from each of 500 nests, which were collected over a period of 20 years. Our goal was to infer kinship between nests based on their genetic relatedness (sharing of same gene sequences).
Felix extracted DNA, constructed genomic libraries for sequencing (ddRAD sequencing) and shipped them off to be sequenced in America. He got a total of 4.4 billion sequencing reads (each only 150 base long) for the 1000 samples. He ran a bioinformatic pipeline that he constructed to process these reads, map them to the reference genome, identify polymorphic sites, and use them to identify genomic segments that are shared between any pair of samples due to their relatedness. We've done the same type of analysis in a previous project on Israeli desert ants and it worked beautifully. To confirm that it works for the American harvester ants, Felix plotted the distribution of kinship scores for all 1000 squared pairs of samples, and colored in red the scores for relatedness between the two ants that were sampled from the same nest (they're assumed to be sisters). This is what he got:

Clearly this is a complete failure. Most of the sisters have relatedness scores that are not bigger than the general distribution of scores between unrelated samples. Felix investigated the data quality and checked (QA) every step of the pipeline. The data quality is very good! He then tried to fine-tune many parameters of different steps in the pipeline and experimented with lots of methodological variations and completely different methods for kinship inference. Nothing works! We think that there is a special challenge in this analysis because of the very unusual genetic structure of this population that is a result of its very weird reproductive system - the so called "dependent lineage system":
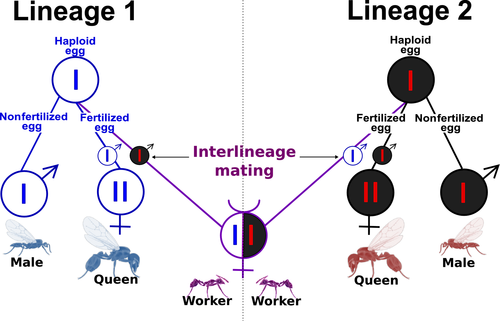
Felix keeps trying multiple variations on the standard pipeline, including some very non-standard methodologies such as a pan-genome approach.
During this project Felix generated a huge, high-quality dataset of 4.4 billion sequencing reads. The unzipped sequence data files take up 1.5TB. The many steps of the processing pipeline and all the variations increase the disk signature many fold. At some point Felix's directory reached 26TB (out of the 45TB total lab quota) before he was lynched by the other lab members...
