- Details
Have you ever failed in something and yet felt very proud? This project is the ultimate example for that. This is the Ph.D. project of Felix Glinka in the Privman lab , which is a collaboration with the lab of Deborah Gordon in Stanford. Deborah's lab has been studying this unique and interesting population of harvester ants in the Arizona desert for almost 40 years. She sent Felix 2 ant samples from each of 500 nests, which were collected over a period of 20 years. Our goal was to infer kinship between nests based on their genetic relatedness (sharing of same gene sequences).
Felix extracted DNA, constructed genomic libraries for sequencing (ddRAD sequencing) and shipped them off to be sequenced in America. He got a total of 4.4 billion sequencing reads (each only 150 base long) for the 1000 samples. He ran a bioinformatic pipeline that he constructed to process these reads, map them to the reference genome, identify polymorphic sites, and use them to identify genomic segments that are shared between any pair of samples due to their relatedness. We've done the same type of analysis in a previous project on Israeli desert ants and it worked beautifully. To confirm that it works for the American harvester ants, Felix plotted the distribution of kinship scores for all 1000 squared pairs of samples, and colored in red the scores for relatedness between the two ants that were sampled from the same nest (they're assumed to be sisters). This is what he got:

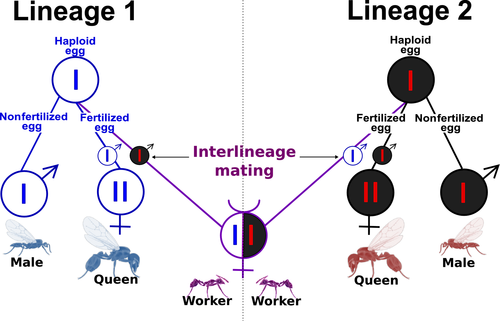
Clearly this is a complete failure. Most of the sisters have relatedness scores that are not bigger than the general distribution of scores between unrelated samples. Felix investigated the data quality and checked (QA) every step of the pipeline. The data quality is very good! He then tried to fine-tune many parameters of different steps in the pipeline and experimented with lots of methodological variations and completely different methods for kinship inference. Nothing works! We think that there is a special challenge in this analysis because of the very unusual genetic structure of this population that is a result of its very weird reproductive system - the so called "dependent lineage system":
Felix keeps trying multiple variations on the standard pipeline, including some very non-standard methodologies such as a pan-genome approach.
During this project Felix generated a huge, high-quality dataset of 4.4 billion sequencing reads. The unzipped sequence data files take up 1.5TB. The many steps of the processing pipeline and all the variations increase the disk signature many fold. At some point Felix's directory reached 26TB (out of the 45TB total lab quota) before he was lynched by the other lab members...
- Details
A new approach to detect and measure echoes in data has been formulated, which is applicable to cases where irregular sampling is concerned.
Proving the feasibility of this approach, required a significant computational effort, which would have taken a normal computer about 10 years to complete. Hive has been instrumental in finishing this project in finite time. The accompanying image shows the results of Monte Carlo simulations that benchmark the ability of the various statistical estimators considered to uncover the input lag (at 100 days). Paper has been submitted to the Astrophysical Journal.
- Details
Dr. Muhammad Akashi who is working with Prof. Doron Chelouche has been running FLASH code models to simulate the formation and evolution of planetary nebulae formed by aging stars.

Link: http://hivehpc.haifa.ac.il/images/dens3D.png
The above picture shows the density structure of the planetary nebula expanding out from three interacting stars. It's the first time such a calculation has been done with such detail, and what we see is a particular frame in time, for a particular projection of the (three-dimensional) nebula.
For comparison, A real planetary nebula, as seen in space is looking like that:

Link(source: www.esa.int ): http://www.esa.int/var/esa/storage/images/esa_multimedia/images/2015/07/born-again_planetary_nebula/15533530-1-eng-GB/Born-again_planetary_nebula.jpg
These results have just been presented in a big meeting on the physics of planetary nebulae by Muhammad in China.
- Details
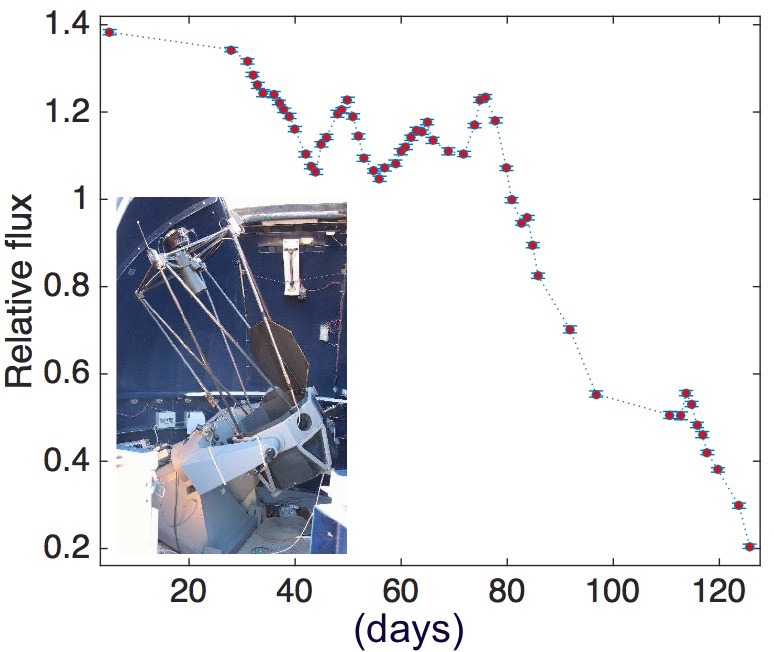
In a first-of-its-kind effort to accurately estimate mass of super-massive black holes, researchers led by Prof. Doron Chelouche, are carrying out nightly photo-metric observations at the Wise Observatory with a follow-up analysis using novel image reduction techniques. This project has far-reaching implications for the formation and evolution of supermassive black holes, the extreme physics in the immediate vicinity of such objects, as well as for the use of quasars as standard cosmological candles.
The outlined study can only be accomplished using a large computer with many processors and a fast distributed file system. Specifically, a normal computer would be unable to cope with the influx of data from the telescope, and the demanding computational task of analyzing them.
The attached image shows the telescope in the inset, and an example for a light curve (i.e., the light-intensity variation over time) for a particular object, obtained by Dr. Francisco Pozo-Nuñez, which harbors a black hole at its center. Several papers are in preparation.
- Details
Tal Yahav from the Privman lab is using the "Queens" of the Hive (high memory servers) to assemble the genome of a new ant species that he himself collected, extracted, and sequenced. The species Cataglyphis drusus is a desert ant native to Israel, which will serve as the model organism for a study in our lab looking for the genetic basis of nestmate recognition in ants.
Tal extracted both DNA and RNA from a single male ant. Genomic DNA sequencing yielded 46 GigaBase (46E9 bases) of sequence data, which Tal assembled into a first draft sequence of the genome. This type of analysis requires very large memory, which is why we purchased the Queens - servers with 768 GB RAM.

- Details
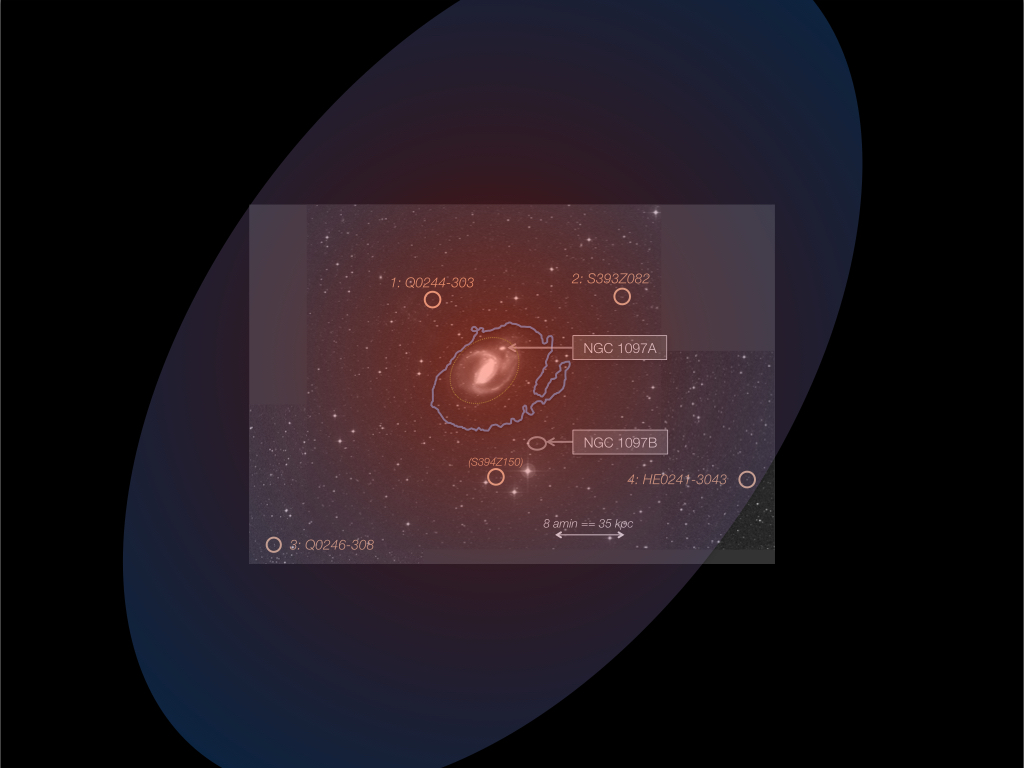
Prof. Doron Chelouche and colleagues from Princeton University have detected, for the first time in the local universe, a gigantic gaseous disk revolving around a galaxy. The size of the disk exceeds the visible part of the galaxy by large factor. This discovery was made possible by acquiring unique spectroscopic data using the Hubble Space Telescope.
Studying the physical properties of this disk has made use of Hive's unique computing capabilities in solving thousands of coupled, non-linear differential equations to uncover the physical state of the gas.
The accompanying image shows the galaxy; the recently detected gaseous component, in the form of a titled disk around it, is marked.
Results have recently been published by the Astrophysical Journal.
- Details
Page 1 of 2